Identifying sections where the element aperture for V6.501 differs from the continuous model for V6.500
Develop the method first with just one aperture component. Build a table
![]()
| LHCB1$START | 0 | 0 | 0.029 |
| IP1 | 0 | 0.029 | 0.029 |
| MBAS.R1 | 2.65 | 0 | 0.029 |
| TAS.1R1 | 20.915 | 0.017 | 0.0193214 |
| BPMSW.1R1.B1 | 21.475 | 0.03 | 0.03 |
| MQXA.1R1 | 29.335 | 0.01895 | 0.01895 |
| MCBX.1R1 | 29.842 | 0.01895 | 0.01895 |
| BPMS.2R1.B1 | 31.529 | 0.0301 | 0.0301 |
| MQXB.A2R1 | 37.55 | 0.024 | 0.024 |
| MCBX.2R1 | 38.019 | 0.024 | 0.024 |
| MQXB.B2R1 | 44.05 | 0.024 | 0.024 |
| TASB.3R1 | 45.942 | 0.027554 | 0.0275543 |
| MQSX.3R1 | 46.7195 | 0.024 | 0.024 |
| MQXA.3R1 | 53.335 | 0.024 | 0.024 |
| MCBX.3R1 | 53.814 | 0.024 | 0.024 |
| MCX3.3R1 | 53.814 | 0.024 | 0.024 |
| MCSOX.3R1 | 54.297 | 0.024 | 0.024 |
| DFBXB.3R1 | 57.972 | 0.0288 | 0.0357966 |
| BPMSY.4R1.B1 | 58.3145 | 0.04 | 0.04 |
| MBXW.A4R1 | 63.022 | 0.064 | 0.064 |

| LHCB1$START | 0 | 0 | 0.029 |
| IP1 | 0 | 0.029 | 0.029 |
| MBAS.R1 | 2.65 | 0 | 0.029 |
| TAS.1R1 | 20.915 | 0.017 | 0.0193214 |
| BPMSW.1R1.B1 | 21.475 | 0.03 | 0.03 |
| MQXA.1R1 | 29.335 | 0.01895 | 0.01895 |
| MCBX.1R1 | 29.842 | 0.01895 | 0.01895 |
| BPMS.2R1.B1 | 31.529 | 0.0301 | 0.0301 |
| MQXB.A2R1 | 37.55 | 0.024 | 0.024 |
| MCBX.2R1 | 38.019 | 0.024 | 0.024 |
| MQXB.B2R1 | 44.05 | 0.024 | 0.024 |
| TASB.3R1 | 45.942 | 0.027554 | 0.0275543 |
| MQSX.3R1 | 46.7195 | 0.024 | 0.024 |
| MQXA.3R1 | 53.335 | 0.024 | 0.024 |
| MCBX.3R1 | 53.814 | 0.024 | 0.024 |
| MCX3.3R1 | 53.814 | 0.024 | 0.024 |
| MCSOX.3R1 | 54.297 | 0.024 | 0.024 |
| DFBXB.3R1 | 57.972 | 0.0288 | 0.0357966 |
| BPMSY.4R1.B1 | 58.3145 | 0.04 | 0.04 |
| MBXW.A4R1 | 63.022 | 0.064 | 0.064 |
| MBXW.B4R1 | 67.288 | 0.064 | 0.064 |
| MBXW.C4R1 | 71.554 | 0.064 | 0.064 |
| MBXW.D4R1 | 75.82 | 0.064 | 0.064 |
| MBXW.E4R1 | 80.086 | 0.064 | 0.064 |
| MBXW.F4R1 | 84.352 | 0.064 | 0.064 |
| X1FCR.4R1 | 141.168 | 0 | 0.02728 |
| BRANA.4R1 | 141.368 | 0 | 0.026 |
| TANAR.4R1 | 142.75 | 0 | 0.026 |
| BPMWB.4R1.B1 | 151.095 | 0.03 | 0.03 |
| MBRC.4R1.B1 | 162.625 | 0.0313 | 0.0313 |
| MCBYV.A4R1.B1 | 164.889 | 0.0289 | 0.0289 |
| MCBYH.4R1.B1 | 166.185 | 0.0289 | 0.0289 |
| MCBYV.B4R1.B1 | 167.481 | 0.0289 | 0.0289 |
| MQY.4R1.B1 | 171.253 | 0.0289 | 0.0289 |
| BPMYA.4R1.B1 | 172.227 | 0.03 | 0.03 |
| TCL.5R1.B1 | 184.857 | 0.04 | 0.04 |
| MCBCH.5R1.B1 | 193.898 | 0.02255 | 0.02255 |
| MQML.5R1.B1 | 198.89 | 0.02255 | 0.02255 |
| BPM.5R1.B1 | 199.635 | 0.024 | 0.024 |
| MCBCV.6R1.B1 | 225.798 | 0.01765 | 0.01765 |
| MQML.6R1.B1 | 230.79 | 0.01765 | 0.01765 |
| BPMR.6R1.B1 | 231.535 | 0.024 | 0.024 |
| XRPV.A7R1.B1 | 237.398 | 0 | 0.0289 |
| BPMSA.7R1.B1 | 237.751 | 0 | 0.0289 |
| XRPV.B7R1.B1 | 241.538 | 0 | 0.0289 |
| DFBAB.7R1.B1 | 258.784 | 0.027633 | 0.0276335 |
| BPM_A.7R1.B1 | 259.259 | 0.024 | 0.024 |
| MQM.A7R1.B1 | 263.404 | 0.022 | 0.022 |
| MQM.B7R1.B1 | 267.171 | 0.022 | 0.022 |
| MCBCH.7R1.B1 | 268.262 | 0.022 | 0.022 |
| S.DS.R1.B1 | 268.904 | 0.022 | 0.022 |
| MCDO.8R1.B1 | 269.245 | 0.022 | 0.022 |
| MB.A8R1.B1 | 283.884 | 0.022 | 0.022 |
| MCS.A8R1.B1 | 284.213 | 0.022 | 0.022 |
| MB.B8R1.B1 | 299.545 | 0.022 | 0.022 |
| MCS.B8R1.B1 | 299.874 | 0.022 | 0.022 |
| BPM.8R1.B1 | 300.698 | 0.024 | 0.024 |
| MQML.8R1.B1 | 306.243 | 0.022 | 0.022 |
| MCBCV.8R1.B1 | 307.335 | 0.022 | 0.022 |
| MCDO.9R1.B1 | 308.311 | 0.022 | 0.022 |
| MB.A9R1.B1 | 322.95 | 0.022 | 0.022 |
| MCS.A9R1.B1 | 323.279 | 0.022 | 0.022 |
| MB.B9R1.B1 | 338.611 | 0.022 | 0.022 |
| MCS.B9R1.B1 | 338.94 | 0.022 | 0.022 |
| BPM.9R1.B1 | 339.765 | 0.024 | 0.024 |
| MQMC.9R1.B1 | 342.941 | 0.022 | 0.022 |
| MQM.9R1.B1 | 346.707 | 0.022 | 0.022 |
| MCBCH.9R1.B1 | 347.798 | 0.022 | 0.022 |
| MCDO.10R1.B1 | 348.777 | 0.022 | 0.022 |
| MB.A10R1.B1 | 363.416 | 0.022 | 0.022 |
| MCS.A10R1.B1 | 363.745 | 0.022 | 0.022 |
| MB.B10R1.B1 | 379.077 | 0.022 | 0.022 |
| MCS.B10R1.B1 | 379.406 | 0.022 | 0.022 |
| BPM.10R1.B1 | 380.23 | 0.024 | 0.024 |
| MQML.10R1.B1 | 385.775 | 0.022 | 0.022 |
| MCBCV.10R1.B1 | 386.867 | 0.022 | 0.022 |
| MCDO.11R1.B1 | 387.843 | 0.022 | 0.022 |
| MB.A11R1.B1 | 402.482 | 0.022 | 0.022 |
| MCS.A11R1.B1 | 402.811 | 0.022 | 0.022 |
| MB.B11R1.B1 | 418.143 | 0.022 | 0.022 |
| MCS.B11R1.B1 | 418.472 | 0.022 | 0.022 |
| BPM.11R1.B1 | 433.013 | 0.024 | 0.024 |
| MQ.11R1.B1 | 437.11 | 0.022 | 0.022 |
| MQTLI.11R1.B1 | 438.579 | 0.022 | 0.022 |
| MS.11R1.B1 | 439.125 | 0.022 | 0.022 |
| MCBH.11R1.B1 | 439.857 | 0.022 | 0.022 |
| S.ARC.12.B1 | 440.285 | 0.022 | 0.022 |
| MCDO.A12R1.B1 | 440.626 | 0.022 | 0.022 |
| MB.A12R1.B1 | 455.265 | 0.022 | 0.022 |
| MCS.A12R1.B1 | 455.594 | 0.022 | 0.022 |
| MB.B12R1.B1 | 470.925 | 0.022 | 0.022 |
| MCS.B12R1.B1 | 471.255 | 0.022 | 0.022 |
| MCDO.B12R1.B1 | 471.947 | 0.022 | 0.022 |
| MB.C12R1.B1 | 486.586 | 0.022 | 0.022 |
| MCS.C12R1.B1 | 486.915 | 0.022 | 0.022 |
| BPM.12R1.B1 | 487.739 | 0.024 | 0.024 |
| MQT.12R1.B1 | 488.653 | 0.022 | 0.022 |
| MQ.12R1.B1 | 492.051 | 0.022 | 0.022 |
| MS.12R1.B1 | 492.581 | 0.022 | 0.022 |
| MCBV.12R1.B1 | 493.313 | 0.022 | 0.022 |
| MB.A13R1.B1 | 508.716 | 0.022 | 0.022 |
| MCS.A13R1.B1 | 509.046 | 0.022 | 0.022 |
| MCDO.13R1.B1 | 509.738 | 0.022 | 0.022 |
| MB.B13R1.B1 | 524.377 | 0.022 | 0.022 |
| MCS.B13R1.B1 | 524.706 | 0.022 | 0.022 |
| MB.C13R1.B1 | 540.037 | 0.022 | 0.022 |
| MCS.C13R1.B1 | 540.367 | 0.022 | 0.022 |
| BPM.13R1.B1 | 541.191 | 0.024 | 0.024 |
| MQT.13R1.B1 | 542.105 | 0.022 | 0.022 |
| MQ.13R1.B1 | 545.503 | 0.022 | 0.022 |
| MS.13R1.B1 | 546.032 | 0.022 | 0.022 |
| MCBH.13R1.B1 | 546.764 | 0.022 | 0.022 |
| E.DS.R1.B1 | 547.188 | 0.022 | 0.022 |
| MCDO.A14R1.B1 | 547.529 | 0.022 | 0.022 |
| MB.A14R1.B1 | 562.168 | 0.022 | 0.022 |
| MCS.A14R1.B1 | 562.497 | 0.022 | 0.022 |
| MB.B14R1.B1 | 577.828 | 0.022 | 0.022 |
| MCS.B14R1.B1 | 578.158 | 0.022 | 0.022 |
| MCDO.B14R1.B1 | 578.85 | 0.022 | 0.022 |
| MB.C14R1.B1 | 593.489 | 0.022 | 0.022 |
| MCS.C14R1.B1 | 593.818 | 0.022 | 0.022 |
| BPM.14R1.B1 | 594.642 | 0.024 | 0.024 |
| MQT.14R1.B1 | 595.556 | 0.022 | 0.022 |
| MQ.14R1.B1 | 598.954 | 0.022 | 0.022 |
| MS.14R1.B1 | 599.484 | 0.022 | 0.022 |
| MCBV.14R1.B1 | 600.216 | 0.022 | 0.022 |
| MB.A15R1.B1 | 615.619 | 0.022 | 0.022 |
| MCS.A15R1.B1 | 615.949 | 0.022 | 0.022 |
| MCDO.15R1.B1 | 616.641 | 0.022 | 0.022 |
| MB.B15R1.B1 | 631.28 | 0.022 | 0.022 |
| MCS.B15R1.B1 | 631.609 | 0.022 | 0.022 |
| MB.C15R1.B1 | 646.94 | 0.022 | 0.022 |
| MCS.C15R1.B1 | 647.27 | 0.022 | 0.022 |
| BPM.15R1.B1 | 648.094 | 0.024 | 0.024 |
| MQT.15R1.B1 | 649.008 | 0.022 | 0.022 |
| MQ.15R1.B1 | 652.406 | 0.022 | 0.022 |
| MS.15R1.B1 | 652.935 | 0.022 | 0.022 |
| MCBH.15R1.B1 | 653.667 | 0.022 | 0.022 |
| MCDO.A16R1.B1 | 654.432 | 0.022 | 0.022 |
| MB.A16R1.B1 | 669.071 | 0.022 | 0.022 |
| MCS.A16R1.B1 | 669.4 | 0.022 | 0.022 |
| MB.B16R1.B1 | 684.731 | 0.022 | 0.022 |
| MCS.B16R1.B1 | 685.061 | 0.022 | 0.022 |
| MCDO.B16R1.B1 | 685.753 | 0.022 | 0.022 |
| MB.C16R1.B1 | 700.392 | 0.022 | 0.022 |
| MCS.C16R1.B1 | 700.721 | 0.022 | 0.022 |
| BPM.16R1.B1 | 701.545 | 0.024 | 0.024 |
| MQT.16R1.B1 | 702.459 | 0.022 | 0.022 |
| MQ.16R1.B1 | 705.857 | 0.022 | 0.022 |
| MS.16R1.B1 | 706.387 | 0.022 | 0.022 |
![]()
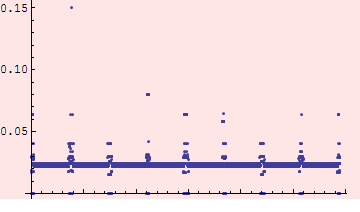
Simple plot of the differences between the two versions of the APER_1 component
![]()
![]()
![]()
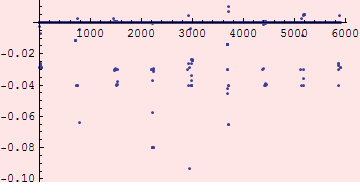
A number of the difference occur because the V6.501 element aperture is unassigned (zero after we processed it) for elements whose names did not appear in the V6.500 sequence. Create a list of elements where the difference is significant but excluding those.
Plotting the s values, we see that they cluster in a few regions.
![]()
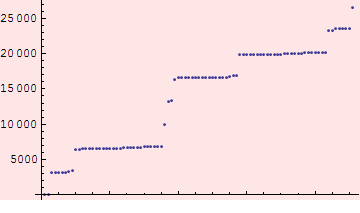
We can try to separate the clusters using a length scale of 200 m
![]()
![]()
![]()
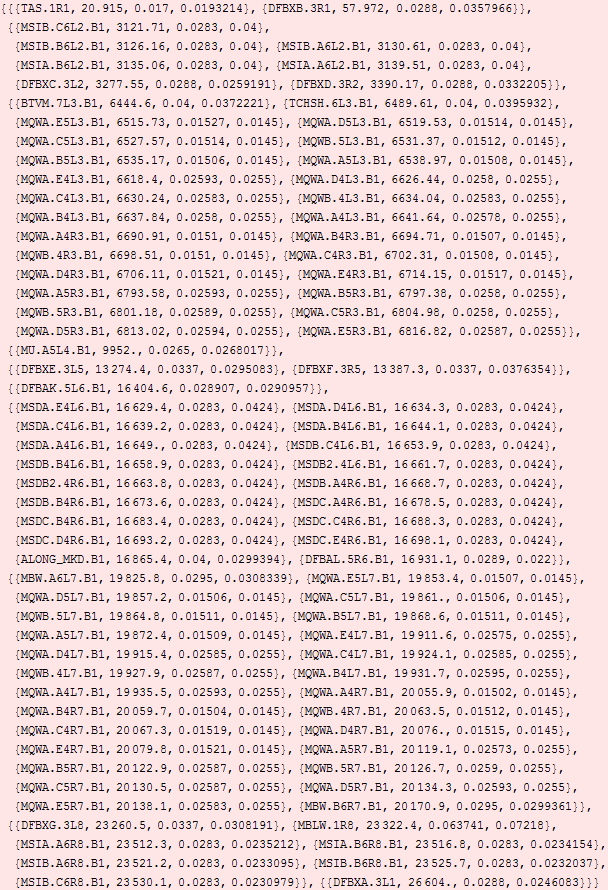
But is that the right scale? Let's look at varying the scale d:
![]()
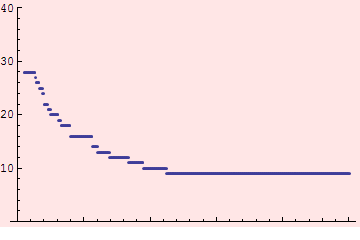
Looks like d=240m would be a good choice. Or maybe 100 m.
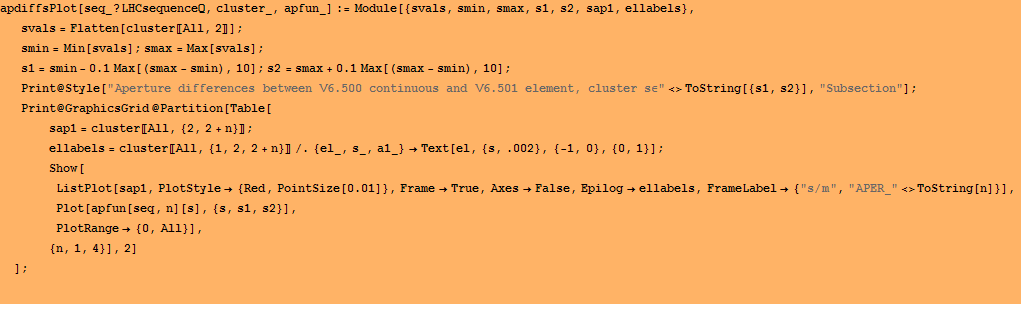
But we need to do this for all 4 aperture components. Anbd then again for LHCB2. Here is a program which defines an array appdiffs[seq] for any sequence and a trial value of the distance scale.

Here we choose our length scale for clustering:

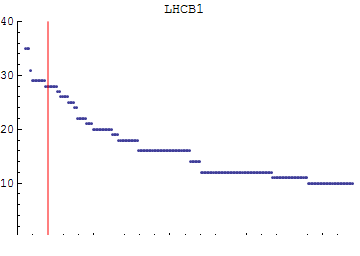
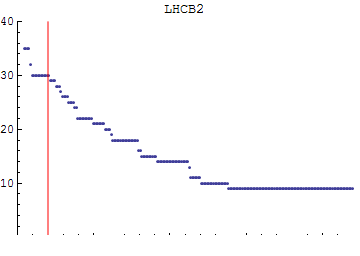
![]()
![]()
![]()
| TAS.1R1 | 20.915 | 0.017 | 0.017 | 0.017 | 0.017 | 0.0193214 | 0.0193214 | 0.0193214 | 0.0193214 |
![]()
![]()
![]()
| Created by Wolfram Mathematica 6.0 (30 August 2007) |